Data Export, R, Matlab, & Python
PAMGuard outputs data both to a SQL relational database and to binary data files.
Users wishing to further process PAMGuard output data in a more flexible programming environment (for example, to develop or run bespoke classifiers on PAMGuard detections, or to make nicer plots for publication), will need to read those data into their programming language of choice.
Most programming languages will be able to connect to and read the database using standard libraries. However, the binary files are in a bespoke format, which requires special software libraries to read data from those files.
Libraries are provided to read data from PAMGuard binary files written in R and MATLAB. A Python library is under development. Read below how to install and use these libraries or go to the full tutorial.
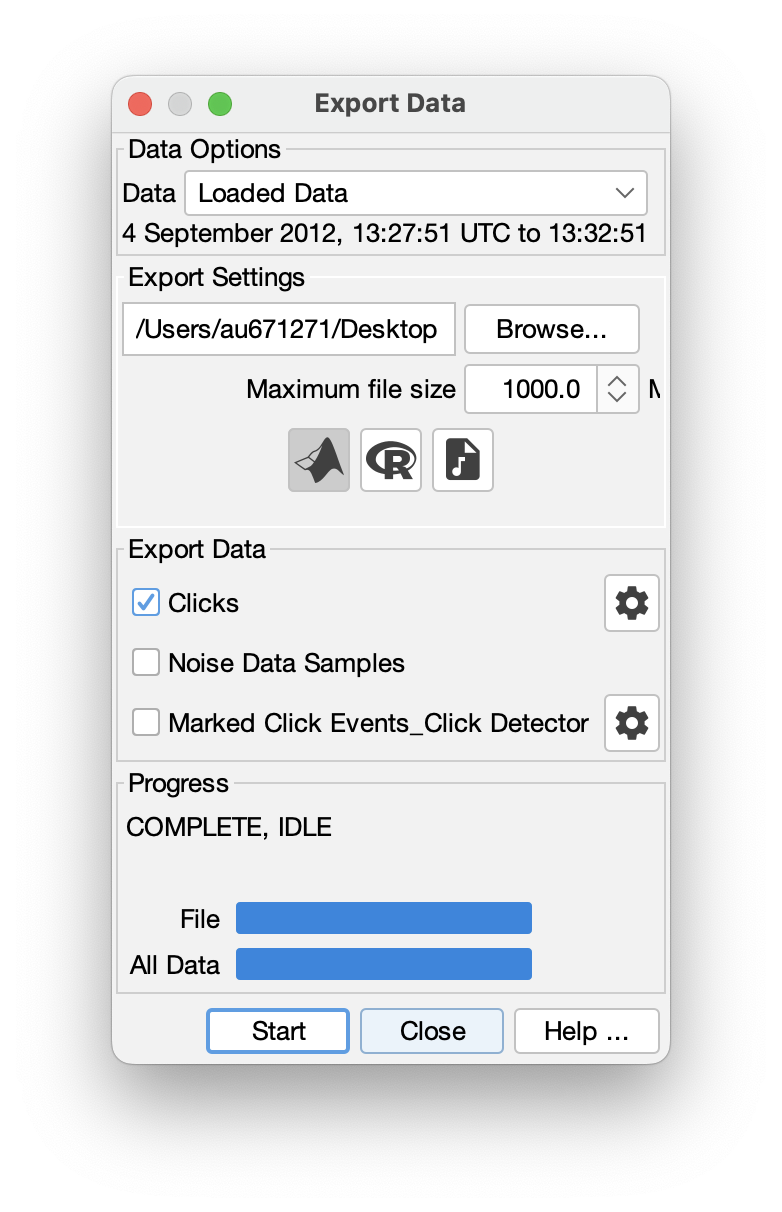
Export data to other platforms
PAMGuard has a powerful in-built exporter which can quickly export detections to;
- .mat files which can be opened in MATLAB.
- .RData frames which can be open in R.
- .wav file which can be opened in any acoustics program.
The exporter allows filtering of data based on, for example, species classification, manual annotations etc. allowing users to specify exactly what is exported.
Thanks to our friends at NOAA Southwest Fisheries Science Center in San Diego, there is a R package for opening PAMGuard binary files:
Installation
You can install the package from CRAN using the following command:
install.packages(“PamBinaries”)
Getting Started
To get started, you can load the package and read in a PAMGuard binary file like so:
library(PamBinaries)
#count the number of classified clicks in a folder of pamguard files.
#folder to a set of binary files.
folder='my/folder/of/pamguard/click/files'
# list all the file names of the
# specified pattern
fnames <- list.files(folder, pattern = "Click_Detector_Clicks.*\\.pgdf$")
count =0; #the classified click counter.
for (val in fnames) {
print(val)
#load each click file.
clickfile <- loadPamguardBinaryFile(file.path(folder, val))
#iterate through the click files to count the classified clicks.
for (aclick in clickfile$data) {
if (aclick$type==1){
count=count+1
}
}
}Links
Users can also take more control and import PAMGuard data files directly in MATLAB using the PAMGuard-matlab library
Installation
MATLAB code for opening PAMGuard binary files is on the PAMGuard GitHub repository.
Download the latest release. Alternatively, you can clone the repository, which will make it easier to get updates if the code is updated.
Getting started
% count the number of classified clicks in a binary file folder of clicks.
clear
% the path to the binary file folder
folder='/my/binary/file/folder';
binaryFiles = dir(fullfile(folder, 'Click_Detector_Clicks*.pgdf'));
count=0;
for i=1:length(binaryFiles)
%print progress
disp(['Loading files: ' num2str(i) ...
' of ' num2str(length(binaryFiles))])
%create filepath from dir dtruct
filePath=fullfile(folder, binaryFiles(i).name);
%load the clicks
clicks=loadPamguardBinaryFile(filePath);
%iterate through all clicks and count how many are classified
for j=1:length(clicks)
if (clicks(j).type==1)
count=count+1;
end
end
endLinks
PAMGuard binary file output can be read in Python using the pypamguard package.
Installation
pip install pypamguardGetting Started
Example of loading in a simple PAMGuard data file into Python.
import pypamguard
df = pypamguard.load_pamguard_binary_file('path/to/data/file.pgdf')Then, for example, you can print out the file header like so.
print(df.file_info.file_header) # File header
print(df.file_info.file_header.version) # File versionModules also have a file_info.module_header, file_info.module_footer, file_info.file_footer, and most importantly data and background.
print("File length", len(df.data)) # Number of module data chunks
first_obj = df.data[0] # First module data chunk
for chunk in df.data: # Looping through the data
print(chunk.identifier)For more information, see the documentation.
Links
Bugs/Requests
Please use the Github issue tracker to submit bugs or feature requests.
License
This software is distributed under the terms of the MIT license. pypamguard is free and open source.
If you use this code, please cite it like so: James Sullivan. (2025). PAMGuard/pypamguard: Version 1.0.0 (v1.0.0). Zenodo. https://doi.org/10.5281/zenodo.16789564
You can find citations for other referencing styles and up-to-date pypamguard versions on Zenodo.