Matched Filter
This method works by cross-correlating a time series input signal with another signal that you specify, called the kernel. Cross-correlation is the optimum method for detecting a known signal in white Gaussian noise. It works best when the signal you wish to detect is quite constant from one instance to the next and from one animal to the next.
In animal bioacoustics it’s actually quite rare that the signal is well enough known (since all natural sounds vary slightly) and the noise background is rarely Gaussian, so this detector is rarely optimal!
Adding the module
From the PAMGuard file menu select Add Modules > Detectors > Ishmael matched filter.
Give a name to the detector, and hit OK.
This detector will require that you also configure a Sound Acquisition module or a Decimator as it’s input.
Configure the module
From the PAMGuard Settings menu select Ishmael matched filter Settings … to open the dialog
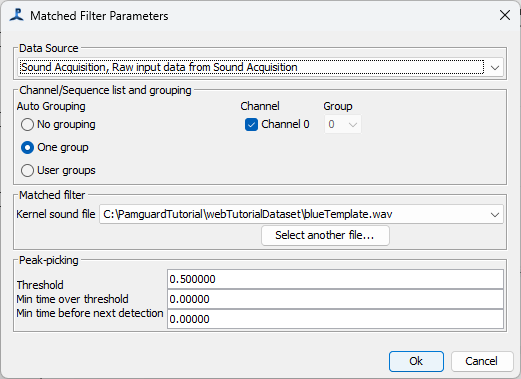
Select the Raw data source. This must be at the same sample rate as the data in the kenel file you’re using with the Matched filter. If necessary, use a Decimator module to get your data to the correct sample rate.
Select which channels you want to process and how you want them to be grouped. If channels are in the same group, it’s slightly easier for the PAMGuard localisers to measure time delays between data on the different channels, but only if the hydrophones in each group are quite close together.
Select the kernel file. This must be a short clip containing a single channel of data that contains a typical example of the sound you’re trying to detect.